#2A Common Proteomics Nextflow Pipeline for Diverse Quality Control Processes: QCloud and QSample
Roger Olivella, Eduard Sabidó, Cristina Chiva, Eva Borràs, Guadalupe Espadas, Olga Pastor, Enrique Alonso, and Selena Fernández
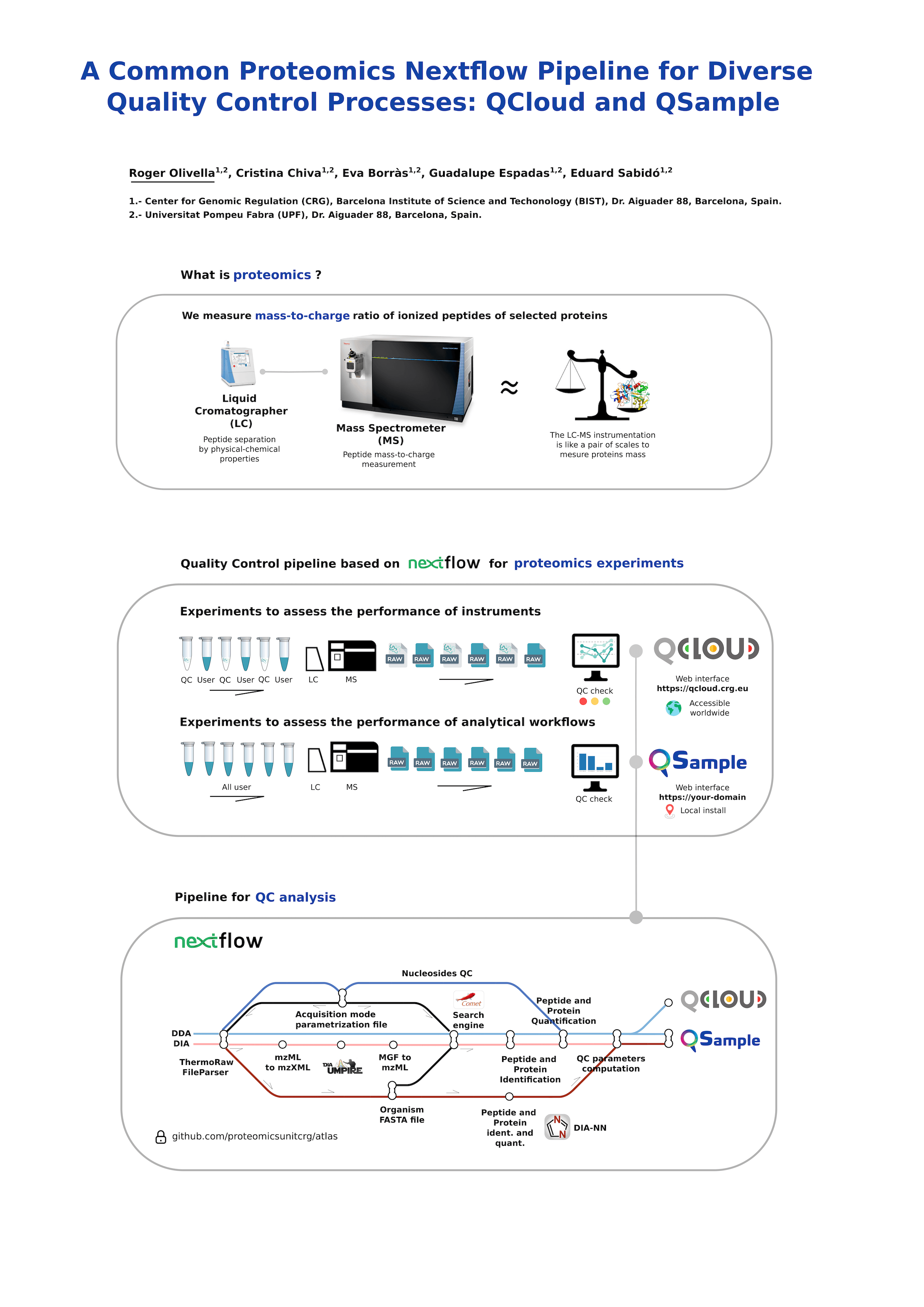
The development of a novel proteomics pipeline capable of integrating with two distinct software tools, QCloud and QSample, is described in this abstract. QCloud is a web-based application designed to evaluate the performance of mass spectrometers, while QSample is another web tool used to assess the quality of proteomics workflows for user samples. The goal was to create a unified pipeline that could seamlessly interface with both software tools, offering a streamlined and efficient workflow for proteomics researchers and unified algorithms for QC parameters computation.
The pipeline employs advanced statistical and bioinformatics methods to perform comprehensive proteomics data analysis. This involves protein identification and quantification. The pipeline combines several state-of-the-art algorithms to facilitate accurate peptide and protein identification, accounting for different variables like post-translational modifications, mass accuracy, database search strategies, etc. to support multiple proteomics applications.