#3FRINGE: Framework for Integrative Genomics
Sander Thierens, Michiel Van Bel, and Klaas Vandepoele
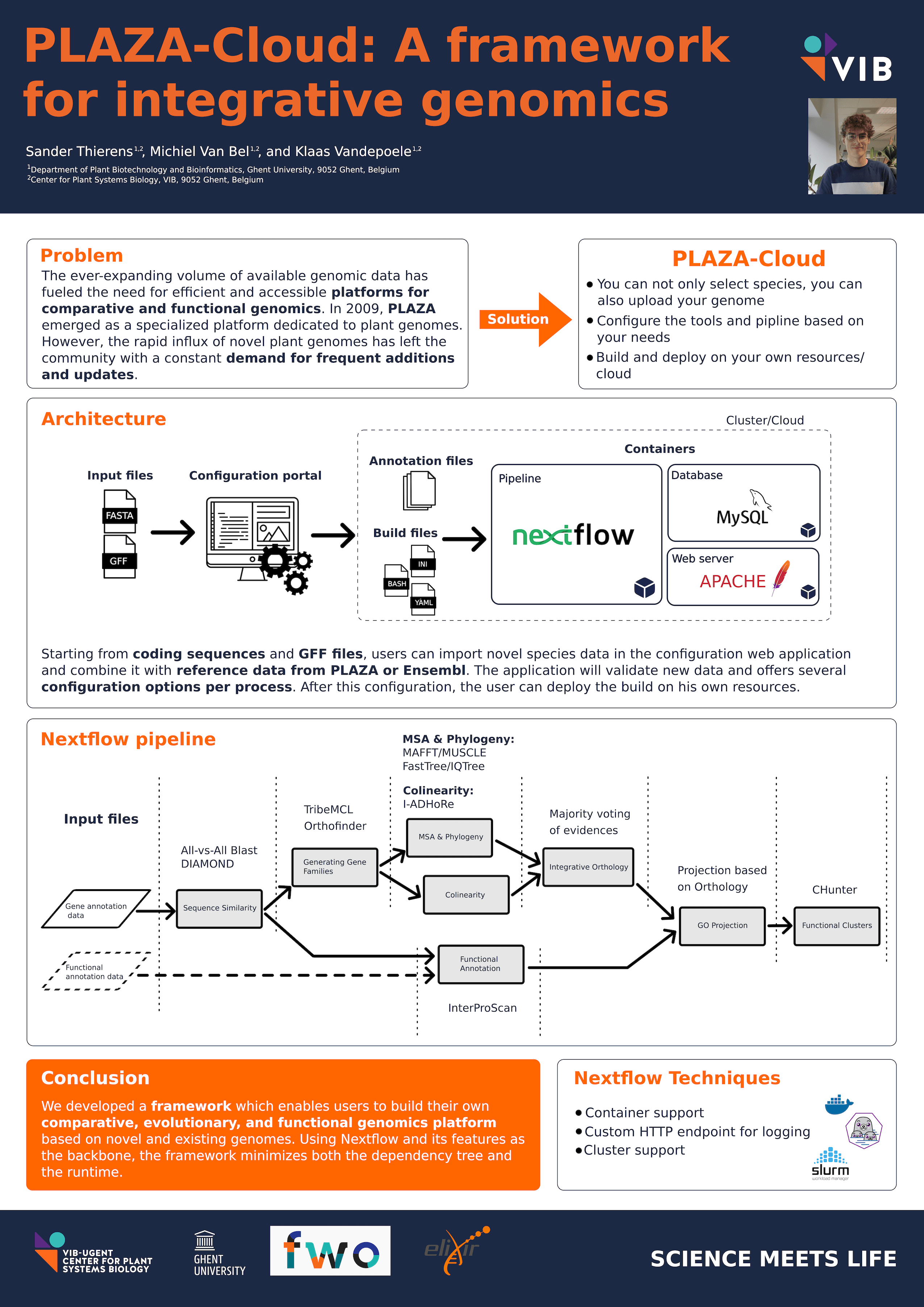
The ever-expanding volume of genomic data generated by numerous genome sequencing initiatives has fueled the need for efficient and accessible platforms for comparative genomics. In 2009, PLAZA emerged as a specialized platform exclusively dedicated to plant genomes and the associated community. However, the rapid influx of novel plant genomes has posed significant challenges, prompting the release of new PLAZA versions every two years. Despite these efforts, the massive growth in new genomes has caused the release schedule to fall behind, leaving the community with a constant demand for more frequent updates and additions.
Addressing these limitations, we present FRINGE (Framework for Integrative Genomics), a novel solution designed to empower researchers by enabling the creation of PLAZA-like environments on their own machines or in the cloud. Through an online application, users can configure customized builds based on newly acquired genomic data or select from a range of provided data sources. This flexible configuration approach initiates the deployment of a Nextflow pipeline, web application, and databases, all contained within virtual environments.
The Nextflow pipeline, integral to this architecture, encompasses various building blocks to derive the necessary data, with each process leveraging the advantages of containerization to further diminish external dependencies. Leveraging Nextflow as the primary pipeline framework provides numerous crucial benefits. These include the ability to cache intermediate result files, enabling users to resume builds after failures, while task statuses are conveniently logged to the online application.
In conclusion, FRINGE accomodates the continuous influx of new genomes without significant resource overhead. With the option to incorporate individual genomic data, it facilitates the creation of builds encompassing data from diverse kingdoms, thereby extending its scope and applicability.