#5Microbiome profiling
Luca Cozzuto, Felipe Morillo, Helene Tonnele, Fernardo Duarte, Julia Ponomarenko, Toni Hermoso Pulido, and Amelie Baud
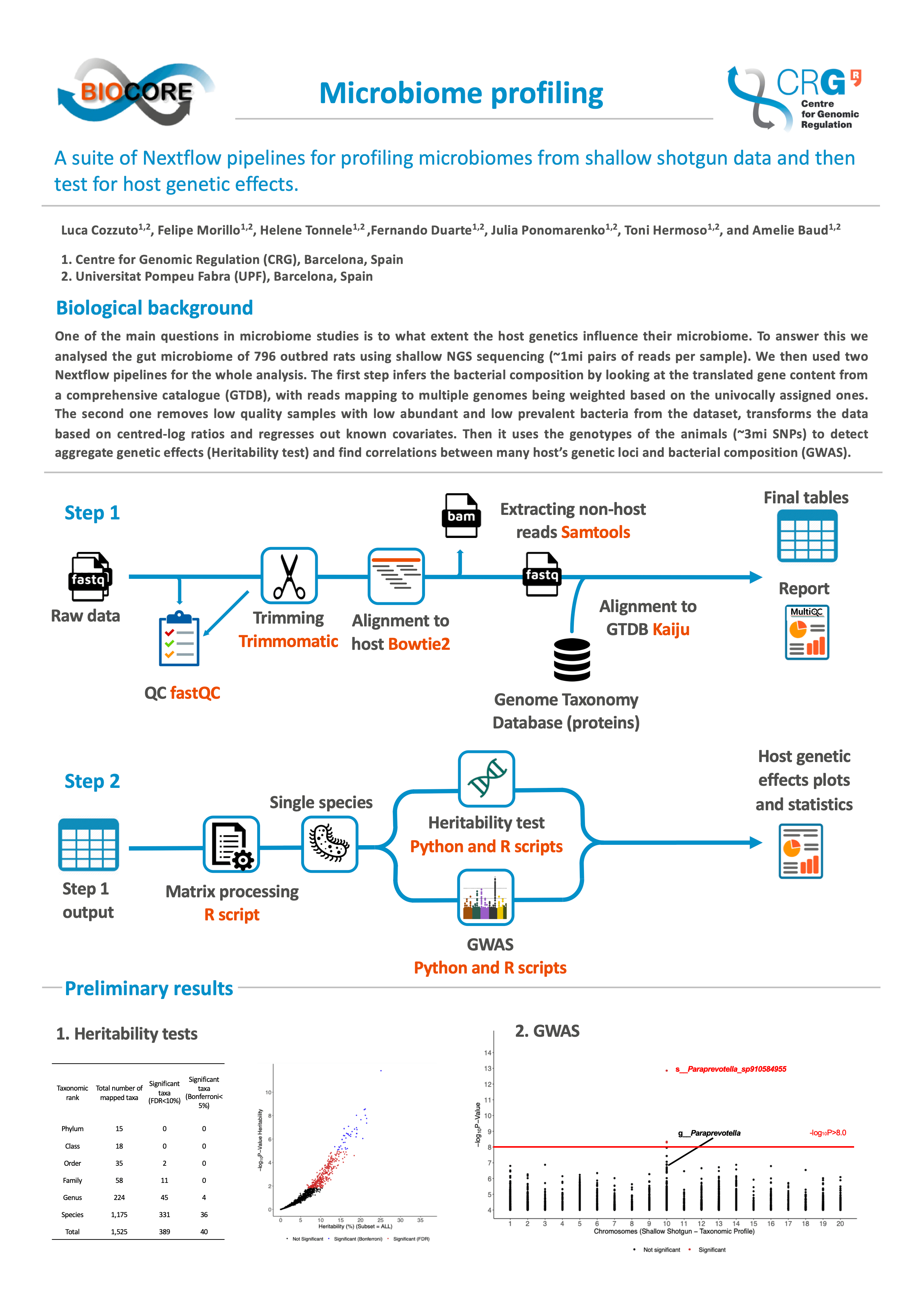
One of the main questions in microbiome studies is to what extent the host genetics influence their microbiome. However, to this date, few variants have been robustly associated with microbiome profiles. We hypothesize this happens because most studies have relied on 16S data, lacking species-level resolution, thus suggesting the adoption of shallow shotgun data, instead. While the main limitation of working with shallow data is the need for good reference catalogues, not all host species have their own catalogue available. Together, working with shallow data and non-conventional hosts bring limitations to using the current pipelines.
We used shallow metagenomic data from the gut of more than 1,000 outbred rats (1 mln pairs of reads per sample) to develop and test a two-step Nextflow pipeline that firstly produces microbiome taxonomic profiles based on a vast bacterial and archaeal genome catalogue, the Genome Taxonomy Database (GTDB). Then, it crosses the resulting profiles with host genotypes to estimate the aggregate heritability of the taxa mapped and perform Genome-Wide Association Studies (GWAS), in order to identify host variants affecting the microbiome. The main advances of this pipeline involve coping with the limitations pointed out above. For example, a DNA-to-protein alignment is applied to increase mapping sensitivity; multiple alignments are proportionally taken into account; low abundance taxa filtering is done on the lowest taxonomic rank before collapsing new count tables for higher ranks and samples with very low coverage are also removed after this. Furthermore, data transformation is done in order to deal with the compositional nature of metagenomic data, covariates are regressed out before the genetic analyses and measurements of alpha and beta diversity are done to provide information about how the microbiome is affected by the same covariates. Plots and statistical tests are also built throughout the pipeline, supporting all final analyses.