#9A multi-viral genome reconstruction pipeline for the integrated molecular surveillance at the RKI
Marie Lataretu, Thomas Krannich, Sofia Paraskevopoulou, and Stephan Fuchs
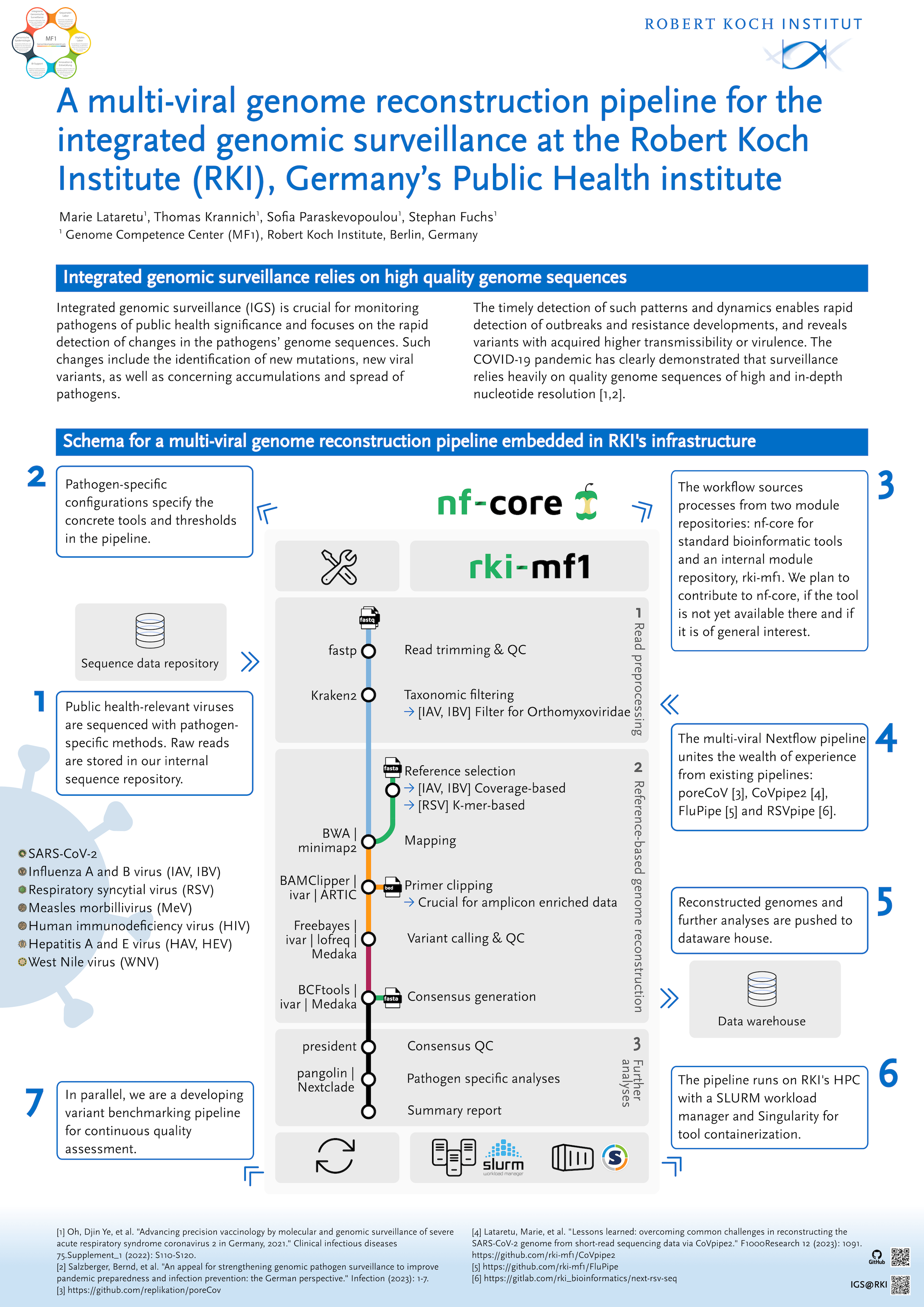
Integrated molecular surveillance is crucial for monitoring pathogens of public health significance and focuses on the rapid detection of changes in the pathogens’ genome sequences. Such changes include the identification of new mutations, new viral variants, as well as concerning accumulations and spread of pathogens. The timely detection of such patterns and dynamics enables rapid detection of outbreaks and resistance developments, and reveals variants with acquired higher transmissibility or virulence. The COVID-19 pandemic has clearly demonstrated that surveillance relies heavily on quality genome sequences of high and in-depth nucleotide resolution.
For this purpose, we are developing a multi-viral genome reconstruction pipeline in nf-core style matching the needs of pathogen-specific sequencing methods and the requirements set by our pathogen experts at the Robert Koch Institute (RKI). The general structure compromises three generic steps (subworkflows): a) read preprocessing and QC, b) reference-based genome reconstruction and consensus generation, and c) further post-reconstruction analysis, e.g., annotation, lineage assignment, outbreak detection, and phylogenetics. Each step is modularized and utilizes pathogen-specific configs defining pathogen-specific tools and parameters. The main workflow uses, on the one hand, nf-core modules and subworkflows for commonly used tools and methods and, on the other hand, an internal module repository for custom scripts and refinements. We plan to contribute to nf-core, if the tool has yet to be available and is of general interest.
The genome reconstruction pipeline is accompanied by a variant benchmarking pipeline for continuous quality assessment and is embedded into a general infrastructure for the integrated genomic surveillance at the RKI.