#18nf-core/molkart: An end-to-end pipeline for the analysis of Molecular Cartography™ data
Kresimir Bestak, Florian Wünnemann, and Denis Schapiro
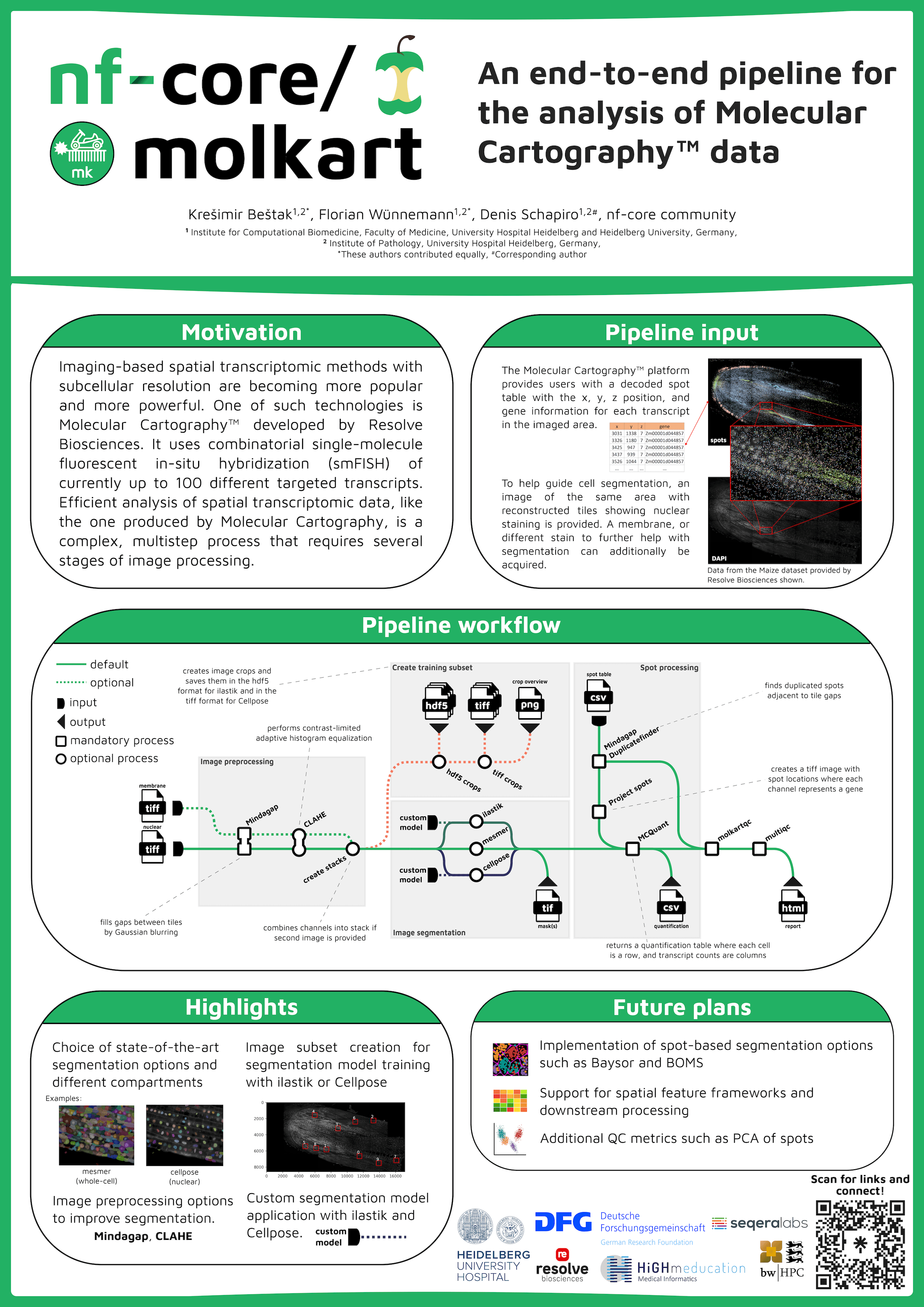
Imaging-based spatial transcriptomic methods with subcellular resolution are becoming more popular and more powerful. One of these technologies is Molecular Cartography™ developed by Resolve Biosciences. It uses combinatorial single-molecule fluorescent in-situ hybridization (smFISH) of currently up to 100 different targeted transcripts. Efficient analysis of spatial transcriptomic data, like the one from Molecular Cartography, is a complex, multistep process that requires several stages of image processing. The data usually consists of a table of locations for single RNA molecules on the tissue as well as images for nuclear and potential membrane staining to help guide cell segmentation. We created molkart, an nf-core pipeline to process Molecular Cartography data using existing nf-core modules from nf-core/mcmicro and new modules tailored to molkart. The pipeline preprocesses nuclear and membrane stains and allows cell segmentation using state-of-the-art methods (e.g., Cellpose 2, ilastik, Mesmer). It also allows the creation of small training data sets for segmentation methods such as Cellpose or ilastik. nf-core/molkart bridges the gap between image processing and spatial transcriptomic analysis, by providing a streamlined pipeline to create cell by transcript matrices that can be used in downstream applications such as nf-core/scflow, Seurat or Scanpy.