#14nf-core/msa: A pipeline to run and systematically evaluate Multiple Sequence Alignment methods.
Luisa Santus
Multiple sequence alignment tools are highly popular modelling methods in bioinformatics. Over the past years, a multitude of MSA methods have been developed, reflecting the significant interest and ongoing advancements in this area.
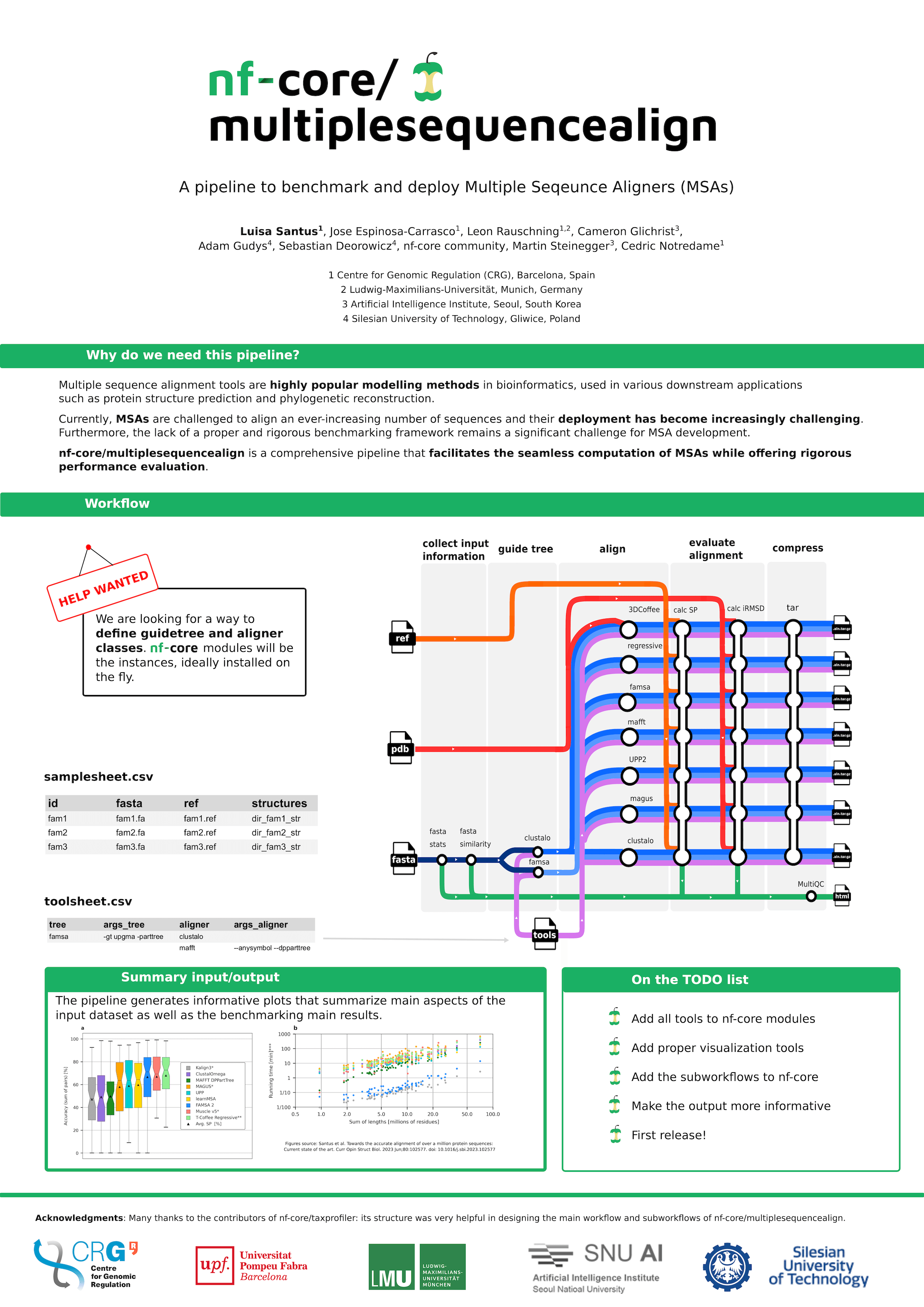
As the demand for MSA methods to align an ever-increasing number of sequences intensifies, their deployment becomes increasingly challenging. The need for a standardized, containerized pipeline to execute such tools in HPC environments has become critical. Furthermore, a major challenge in the MSA development field is the lack of a proper and rigorous benchmarking framework. A standard platform is essential for evaluating tools’ performance systematically and impartially, aiding researchers in making informed decisions when choosing or developing new tools.
To address these challenges, we present a comprehensive pipeline that facilitates the seamless computation of multiple sequence alignments (MSAs) while offering rigorous performance evaluation. The pipeline incorporates the calculation of various input file metrics and evaluation metrics for each executed tool.
The pipeline’s modular design, thanks to Nextflow’s DSL2 extension, allows for the effortless integration of newly developed methods into the benchmark. Moreover, it enables the estimation of the individual contributions of different MSA tool components, such as guide tree rendering, to the final performance metrics.
We envision that the implementation of this pipeline will significantly impact both the MSA development community and its user base. Additionally, its structure can serve as a valuable template for benchmarking other tools as well, extending its potential benefits to various areas of research.