#17Scalepopgen: bioinformatics workflow resources for population genomics
Maulik Upadhyay, Neza Pogorevc and Ivica Medugorac
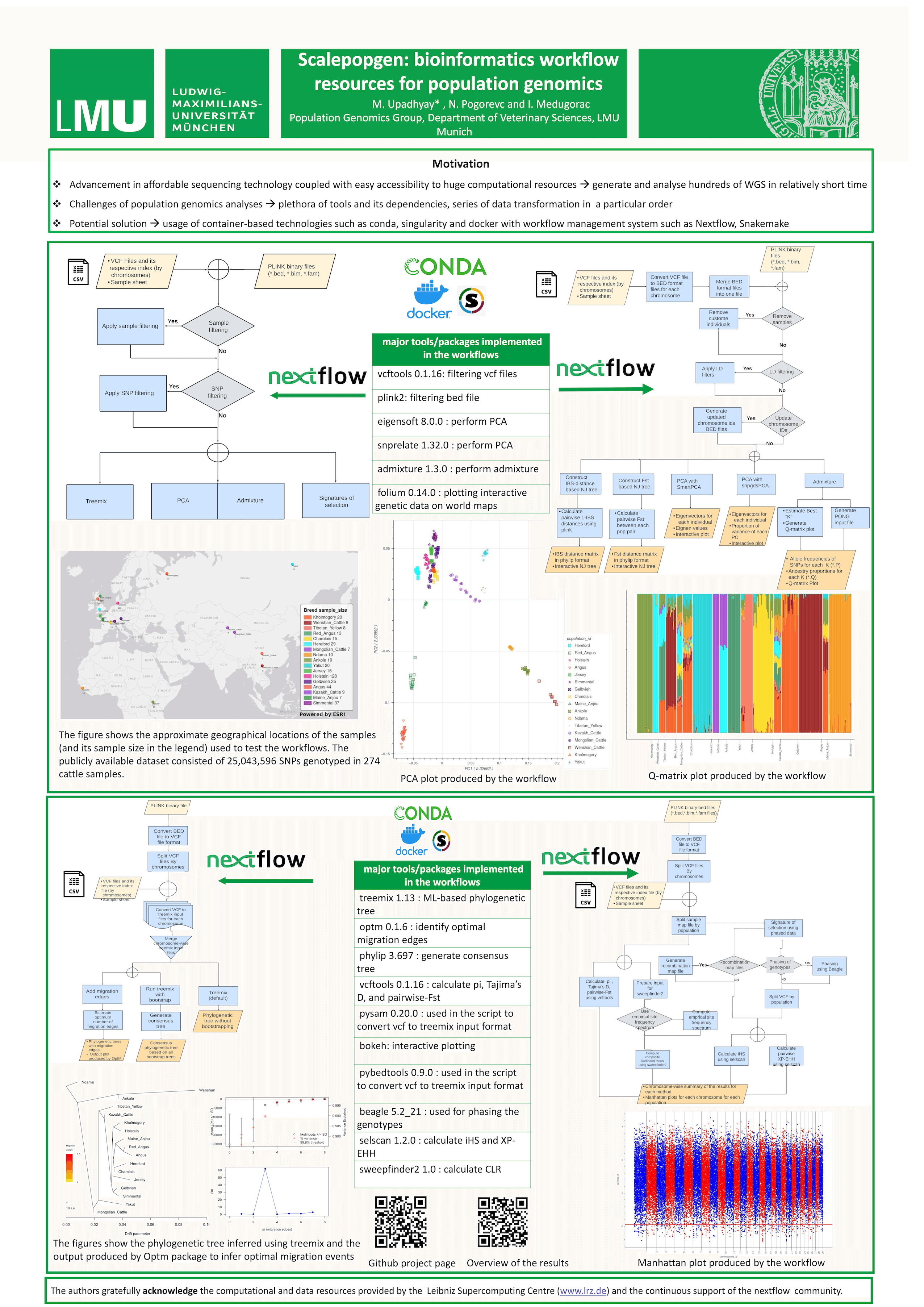
Population genomic analyses such as inference of population structure, detecting introgression, identifying signatures of selection usually involve application of plethora of tools and approaches. Further, installation of tools and dependencies, data transformation or series of data preprocessing in a particular order, sometimes makes population data analyzes challenging. While usage of container-based technologies have significantly resolved the problems associated with installation of tools and its dependencies, population genomics analyses requiring multi-steps pipeline or complex data transformation can greatly be facilitated by application of workflow management systems (WMSs) such as nextflow and snakemake. The usage of such WMSs in population genomics analysis can also contribute to scalability and reproducibility. Here, we describe scalepopgen, a fully automated workflow that can apply widely used population genomics techniques starting from vcf or plink generated binary files. The workflow is developed in nextflow and can be run locally or on high performance computing systems using either conda, singularity or docker. The automated workflow include procedures such as: (1) filtering of individuals and genotypes, (2) running PCA, admixture and identifying optimal K-values, (3) running treemix with or without migration edges, and bootstrapping, followed by identification of optimal number of migration edges, (4) implementing single-population and multi-population comparison-based procedures to identify signatures of selection. The workflow uses various open-source tools as well as several python R scripts, and a cpp tool developed here to implement techniques for which, to our knowledge, no tools were available. The scalability and reproducibility of this pipeline are demonstrated using vcf files generated from hundreds of available livestock genome sequences.