
#7Tomte a pipeline for RNA-seq analysis in rare disease diagnostics
Lucía Peña-Pérez, Anders Jemt, Jesper Eisfeldt, Ramprasad Neethiraj, Esmee ten Berk de Boer, Mei Wu, Nicole Lesko, Anna Wedell, Anna Lindstrand, Valtteri Wirta, and Henrik Stranneheim
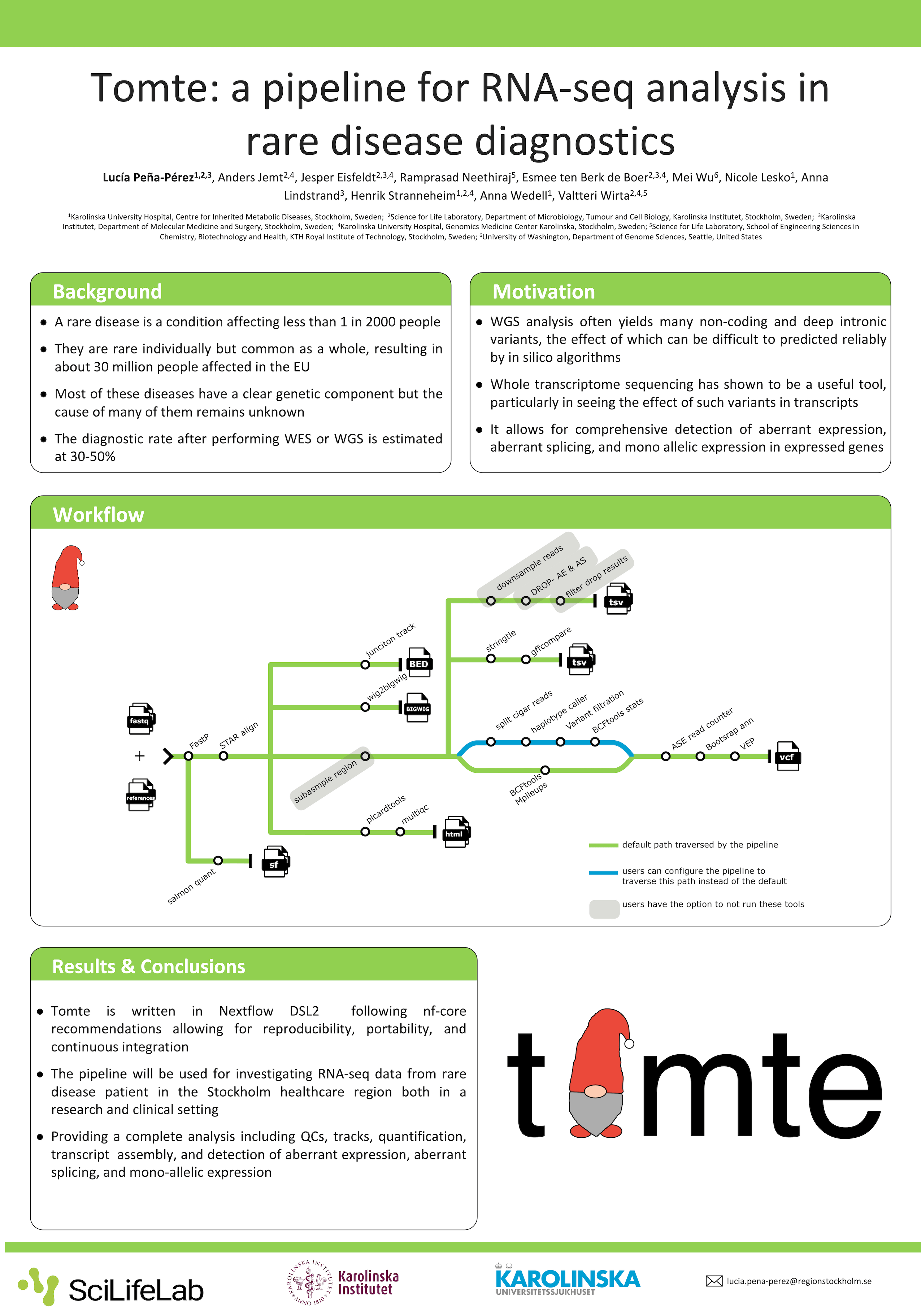
Whole genome sequencing is one of the most comprehensive and broadly used techniques for rare-disease diagnosis. However, many patients remain undiagnosed even after having their genome analyzed. One of the main reasons is the bottleneck in variant interpretation, often caused by the inability to predict the effects of non-coding/deep intronic variants on protein. Performing RNA-seq can allow us to see the effects directly on the transcripts, potentially increasing the diagnostic yield. Herein we describe the development of tomte, a pipeline to analyze RNA-seq data from rare disease patients.
Tomte is a pipeline written in Nextflow adhering to the nf-core guidelines that performs alignment of FASTQ files, transcript quantification, allele-specific variant calling, and variant annotation. Furthermore, it detects mono-allelic expression, aberrant expression, and aberrant splicing to aid in rare disease diagnostics.
The pipeline will be used to analyze RNA-seq data from accessible tissues such as blood, fibroblasts, or muscle of patients with endocrine and metabolic rare diseases in the Stockholm healthcare region. By leveraging the nf-core framework we are able to utilize the standardized tools and modules to create a modular pipeline all while ensuring a simple installation, efficient run, and reproducible results.
Tomte a pipeline for RNA-seq analysis in rare disease diagnostics
