#19The ENA SARS-CoV-2 Drag and Drop Uploader
Zahra Waheed and Ahmad Zyoud
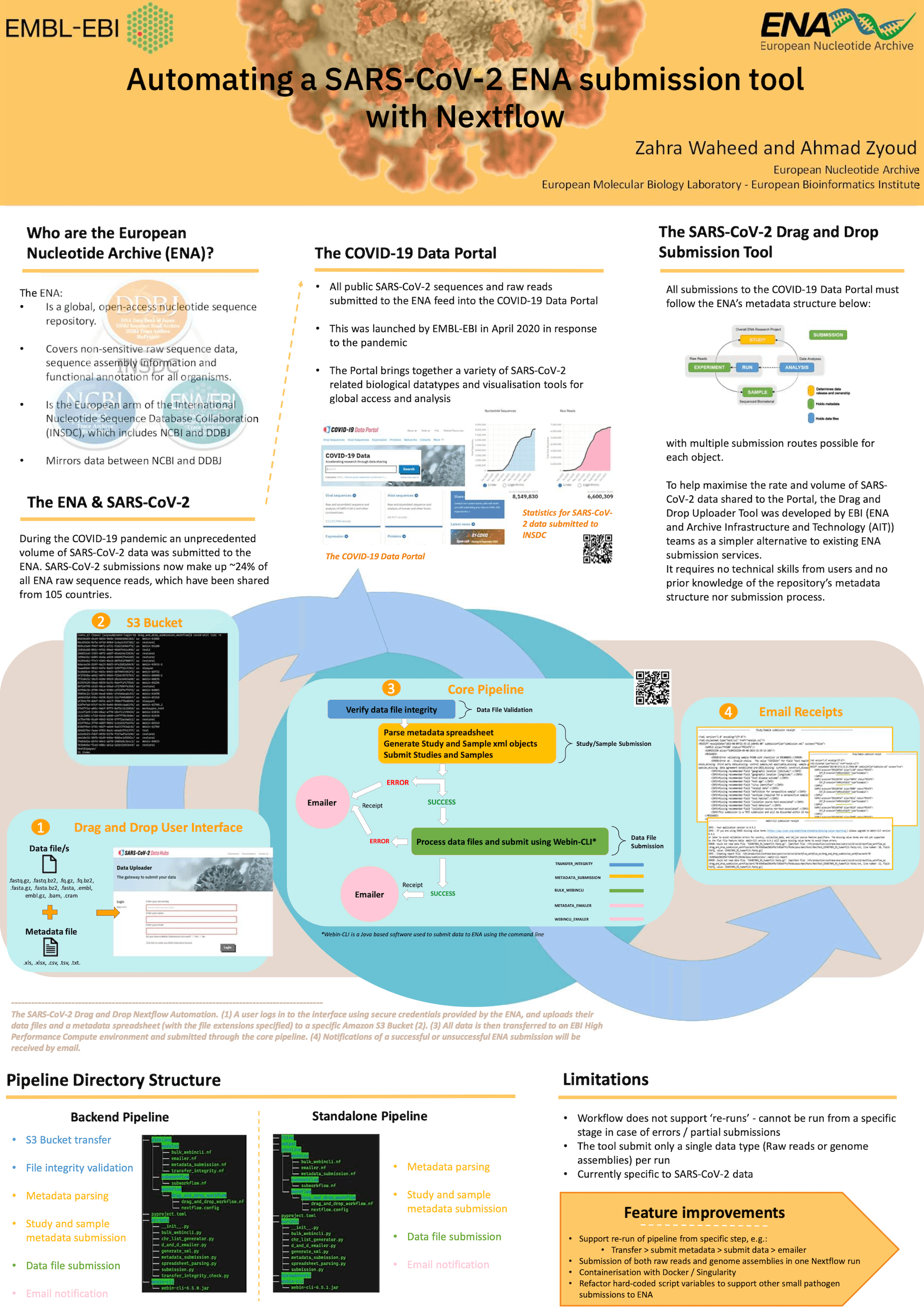
The COVID-19 pandemic highlighted the importance of sharing genomic data and metadata globally, through submission to public nucleotide sequence repositories. To date, over 12 million raw reads and assembled SARS-CoV-2 sequences have been submitted to the European Nucleotide Archive (ENA) alone, which are visible and retrievable from the COVID-19 Data Portal. The rapid sharing of this data (whilst keeping in line with recommended metadata standards) is key to efficient outbreak surveillance and sequence interpretation, and helps to drive a more effective public health response. Thus developing simple, user-friendly submission tools is valuable for lowering the barrier to data entry, and maximising the rate and volume of data shared for scientific research.
Seeing the need for a one-stop shop SARS-CoV-2 submission tool, the ENA team developed the SARS-CoV-2 Drag and Drop Uploader, which requires no technical skills from users and no prior knowledge of the repository’s submission process. The tool offers an alternative route to submit SARS-CoV-2 data to the ENA but can also be repurposed for other viral submissions, as proven by the equivalent ENA Monkeypox Uploader in response to the 2022 outbreak.
Here we present a full Nextflow pipeline for the back-end automation of the SARS-CoV-2 Drop Uploader, incorporating existing ENA APIs, command line programs and AWS.
We additionally present a portable, standalone version of this workflow for ENA data submission.