#15CRISPR gene editing analysis with nf-core/crisprseq: A pipeline for targeted and screening experiments
Júlia Mir Pedrol, Laurence Kuhlburger, Marta Sanvicente-García, Gisela Gabernet, nf-core community, Marc Güell, Ghazaleh Tabatabai, and Sven Nahnsen
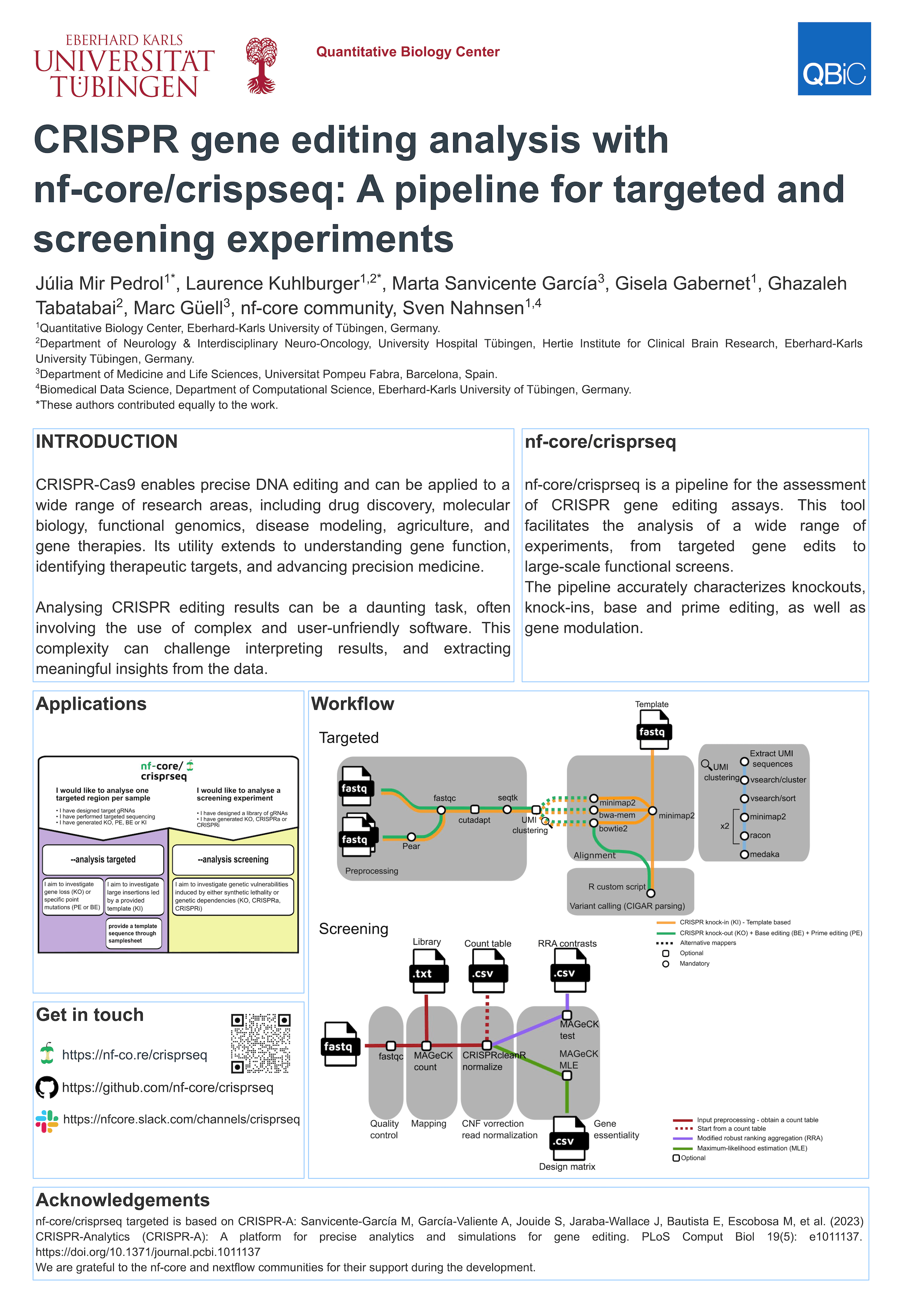
CRISPR-Cas9 gene editing has revolutionized the fields of molecular biology and functional genomics. This technique enables precise DNA editing which unlocks numerous possibilities for understanding gene function as well as identifying therapeutic targets, and advancing precision medicine. It holds big potential in various research domains, including drug discovery, disease modelling, agriculture or gene therapies.
Here we present nf-core/crisprseq, an analysis pipeline for the analysis of CRISPR gene editing experiments. This tool facilitates the analysis of a wide range of experiments, from targeted gene edits to large-scale functional screens. The pipeline accurately characterizes knockouts, knock-ins, base and prime editing, as well as gene modulation, while providing precise editing efficiency assessments and identifying genetic dependencies in screening studies. Written with Nextflow’s DSL2 and incorporating nf-core modules, the pipeline ensures reproducibility and modularity while adhering to community standards.

Júlia Mir Pedrol
Bioinformatician at Quantitative Biology Center - University of Tübingen