#9An Overview of Bioinformatic pipelines for metabarcoding data analyses
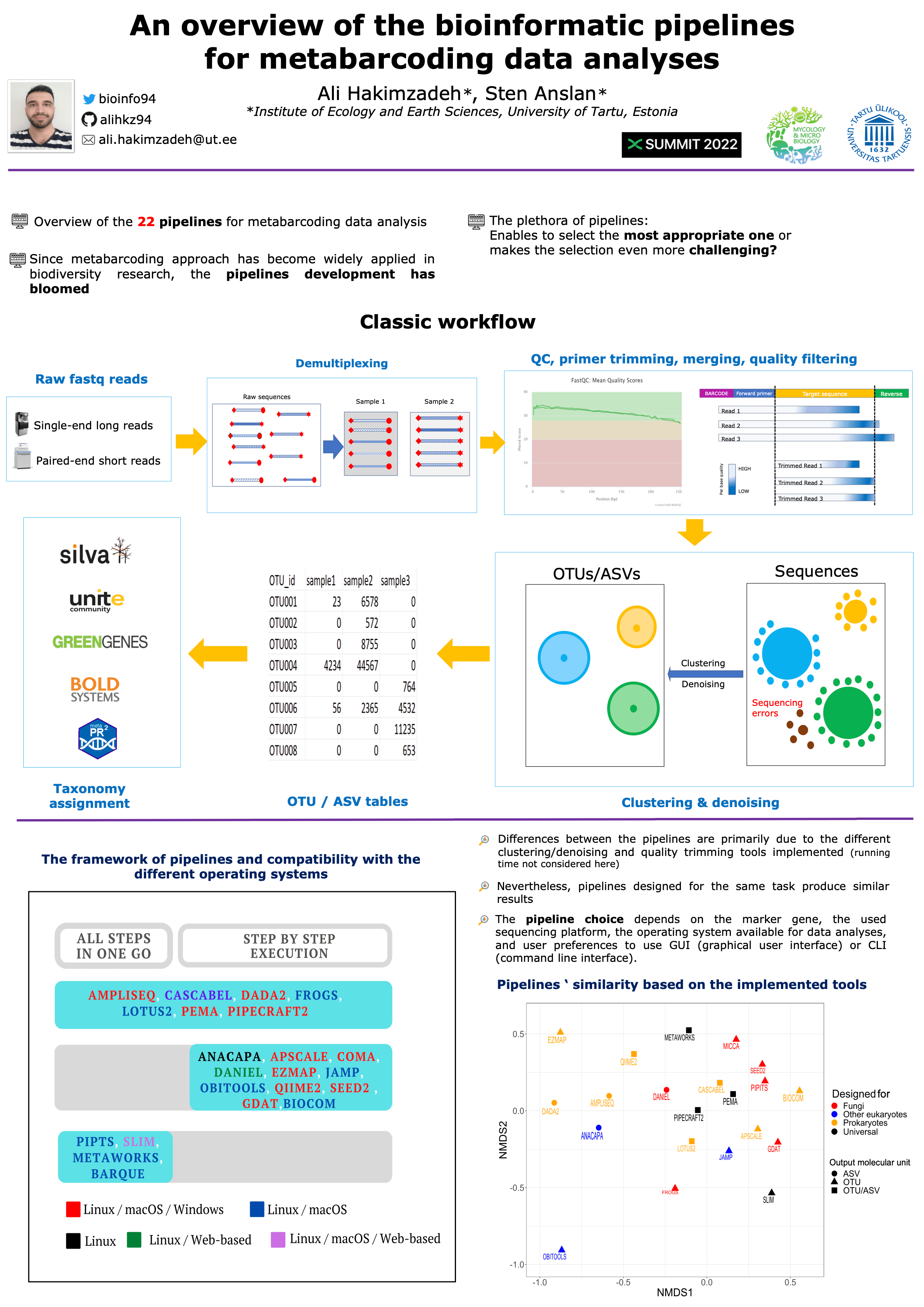
Bioinformatic analysis of eDNA metabarcoding data is a critical component of a comprehensive biodiversity assessment. For community analysis, high-throughput data needs to be processed in several steps, from quality filtering to taxonomy identification. Software suites combine many of these applications to help researchers automate the analysis of the data they collect. Among them, the pipelines most commonly used are QIIME2, LOTUS, and USEARCH. Several pipelines have been developed for COI and ITS marker genes, including BARQUE, FROGS, and PIPITS. Many Pipelines are now available for the required analyses, but their relative abilities to provide fast and accurate species lists have seldom been evaluated. In this review, we outline their specific properties and input data requirements, which aids in navigating among the plethora of amplicon processing pipelines to simplify the software selection for metabarcoding projects.