#3atlas: a novel Nextflow quality control pipeline for a plethora of mass spectrometry workflows
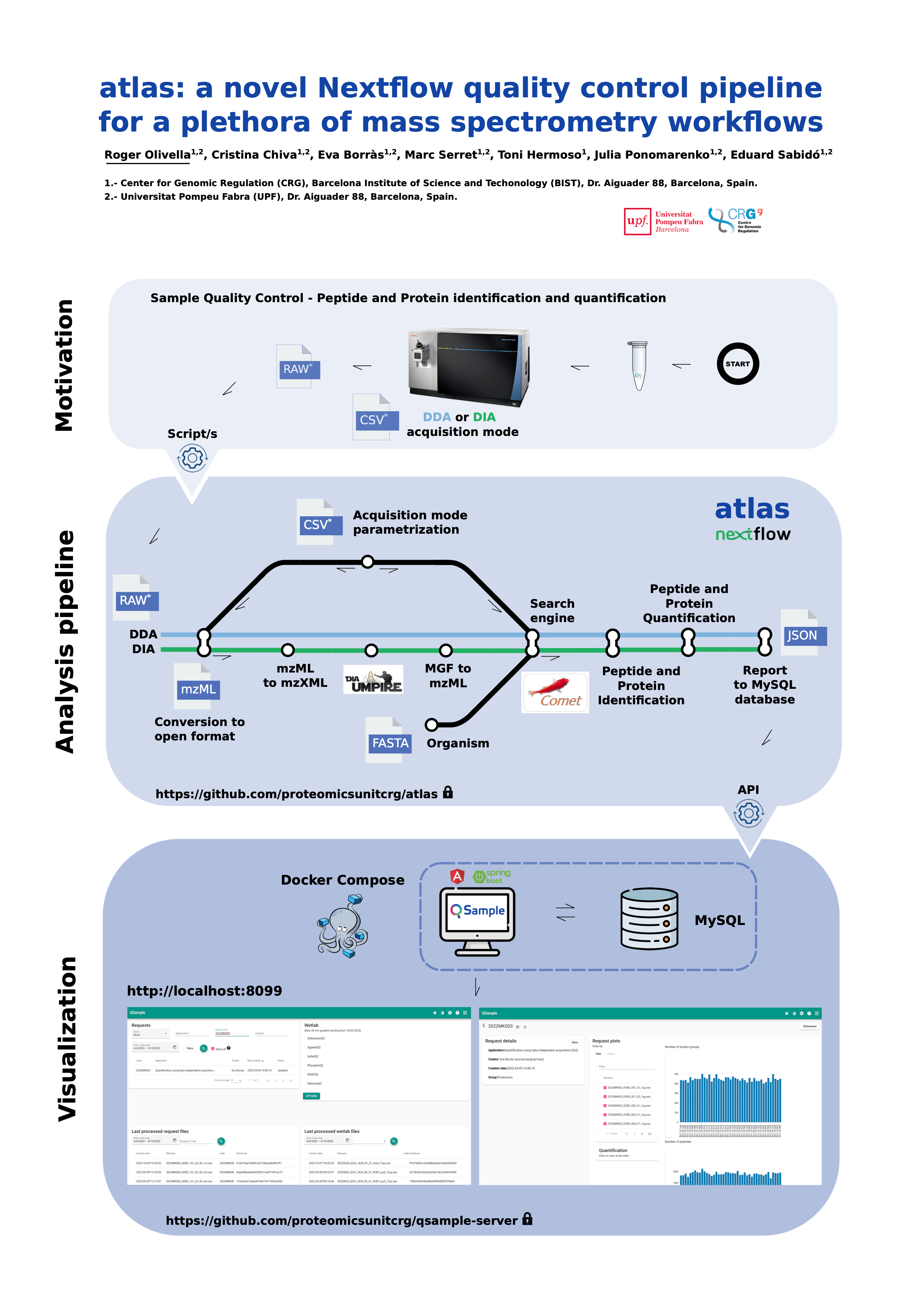
“atlas” is a new Nextflow-based pipeline for processing and analysis of mass spectrometry data. Specifically, atlas is designed to support proteomics laboratories in the daily quality assessment of different proteomics applications. It can work for many kinds of workflows, including the analysis of regular proteomes, phosphoproteomes, quantification of histones and it can process data on independent acquisition mode, among others. For this plethora of workflows, “atlas” is capable of extracting several key quality control parameters that substantially simplify the assessment of the experiments performed on mass spectrometers. “atlas” is based on OpenMS modules, an open source processing software for mass spectrometry data which is containerized using Singularity, a software used to encapsulate all required dependencies.
atlas: a novel Nextflow quality control pipeline for a plethora of mass spectrometry workflows
