#7nf-core/funcscan: Identifying natural products and functional gene clusters in ancient microbial DNA
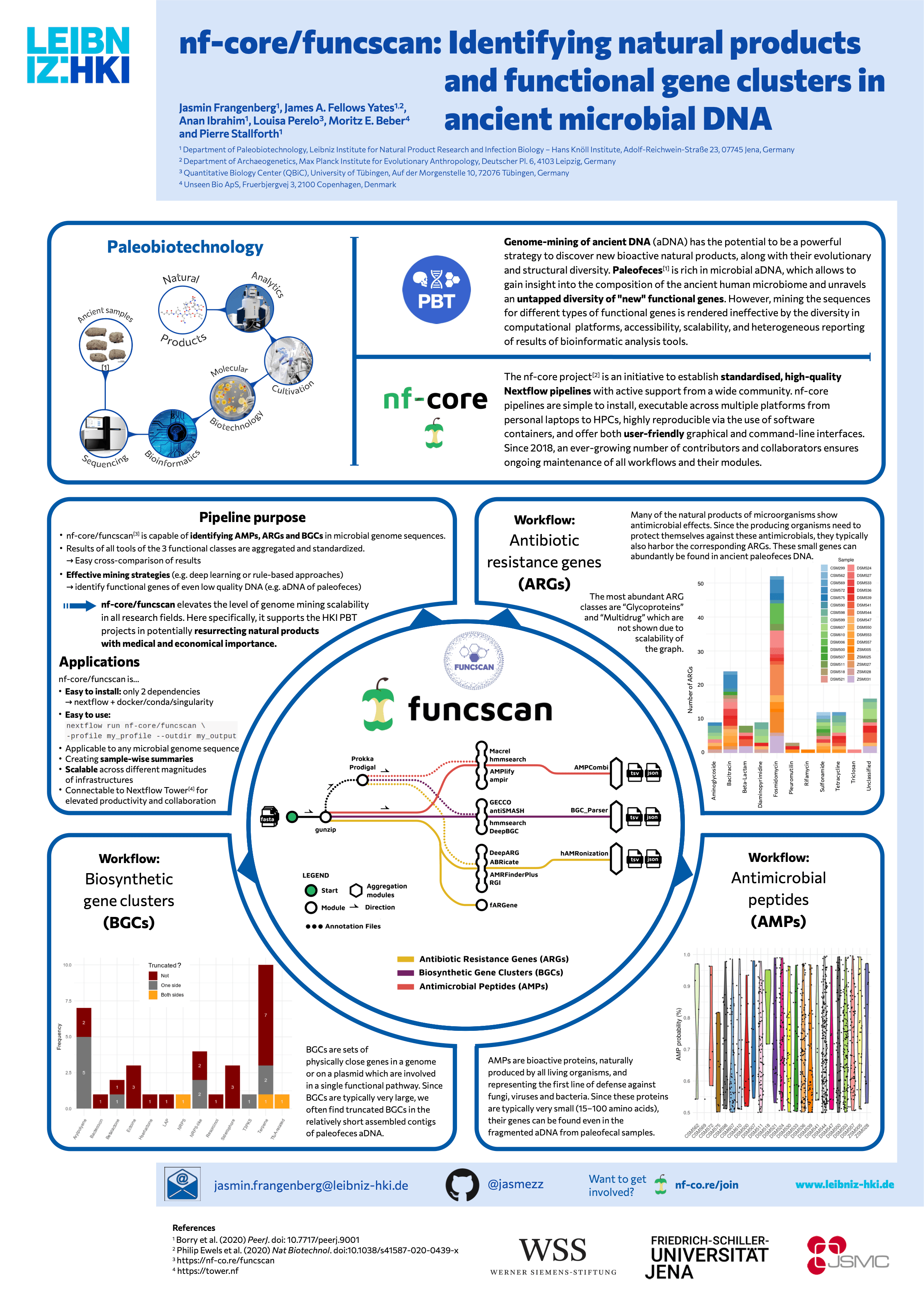
Genome-mining of ancient DNA (aDNA) is a powerful strategy to discover new bioactive natural products, along with their evolutionary and structural diversity. However, mining the sequences is often hampered by the diversity in computational platforms, accessibility, scalability, and heterogeneous result reports of bioinformatic tools. Here, we present nf-core/funcscan, a Nextflow pipeline for screening assembled contigs for functional and biosynthetic components. We demonstrate this functionality using an aDNA dataset from paleofeces. nf-core/funcscan currently provides 12 tools to simultaneously predict antimicrobial peptides, antibiotic resistance genes, and biosynthetic gene clusters from full or partial genomes. The pipeline provides standardization of the outputs, which facilitates the rapid evaluation and visualization of the results. Thus, nf-core/funcscan allows researchers of different scientific fields and backgrounds to efficiently screen genomic data for functional genes.

Jasmin Frangenberg
Doctoral Researcher at the Leibniz Institute for Natural Product Research