#2nf-core/proteinfold: a bioinformatics best-practice pipeline for protein 3D structure prediction
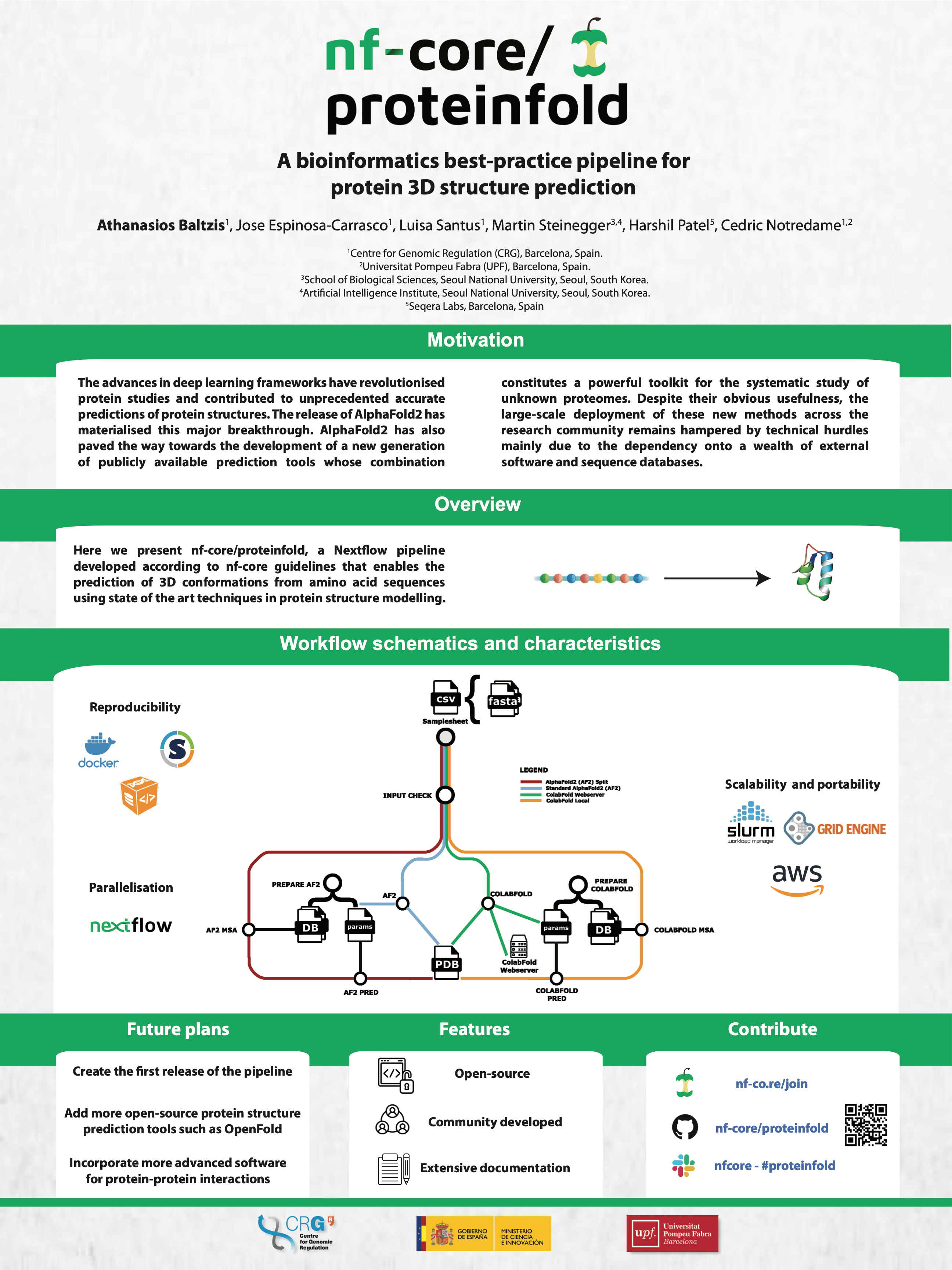
The advances in deep learning frameworks have contributed to unprecedented accurate predictions of protein structures. The release of AlphaFold2 has materialised this major breakthrough and paved the way towards the development of a new generation of publicly available prediction tools whose combination constitutes a powerful toolkit for the systematic study of unknown proteomes. Despite their usefulness, the large-scale deployment of these new methods across the research community remains hampered by technical hurdles. The main challenge is the dependency of each prediction method onto a wealth of software and databases. Here we present nf-core/proteinfold, a Nextflow pipeline developed according to nf-core guidelines that enables the use of state of the art techniques in protein structure modelling. These best-practice guidelines ensure that the pipeline is scalable, reproducible and portable for execution on the major cloud providers as well as HPC infrastructures.