#4Enhancing pathogen surveillance of Candida auris using MycoSNP and Nextflow: an update
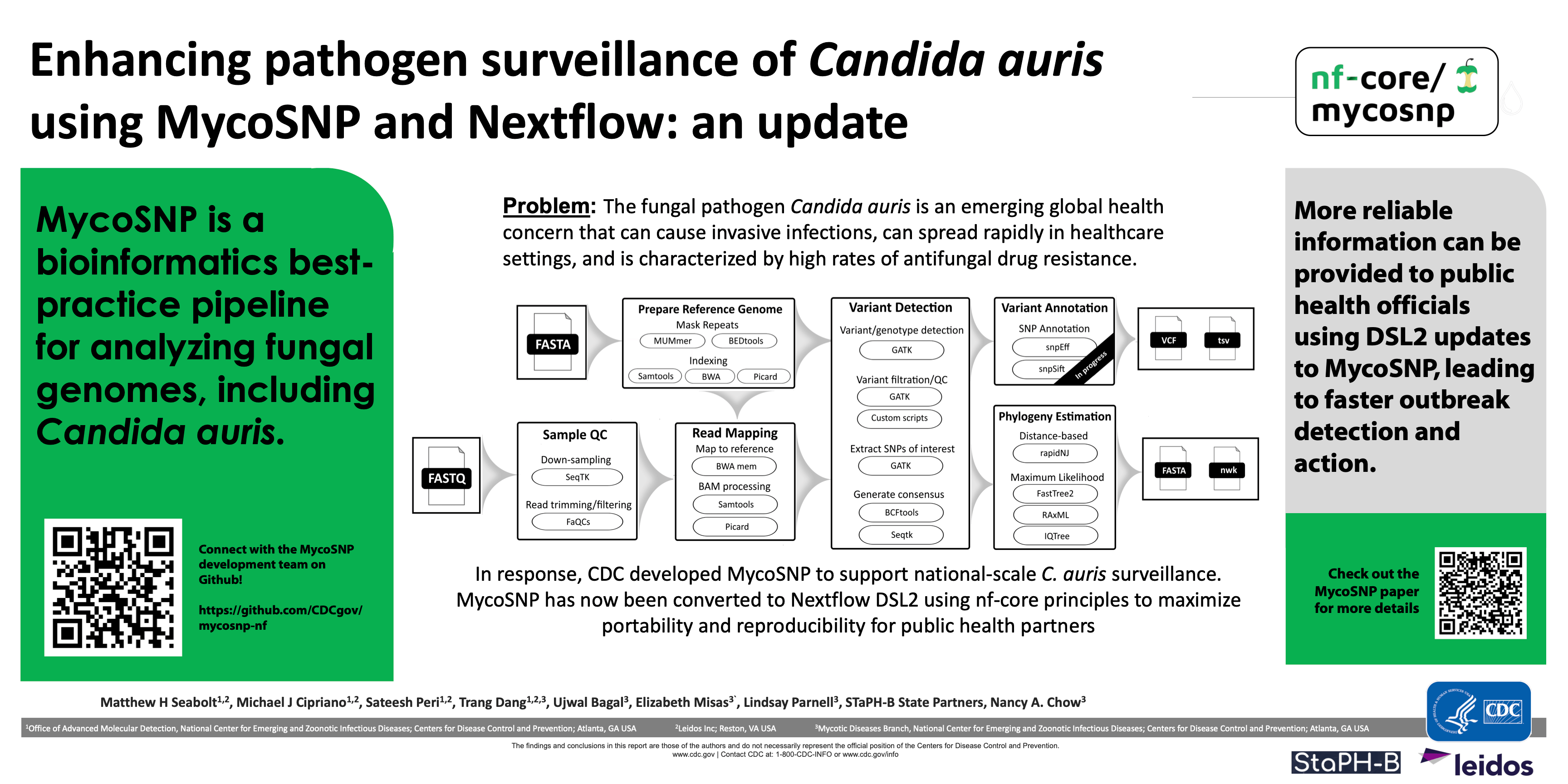
The fungal pathogen Candida auris is an emerging global health concern that can cause invasive infections, can spread rapidly in healthcare settings, and is characterized by high rates of antifungal drug resistance. In response, CDC developed MycoSNP, a portable bioinformatics pipeline for performing whole-genome sequencing analysis of fungal pathogens including C. auris. In brief, MycoSNP ingests raw short-read sequencing data, performs quality control, maps reads to a reference genome, and calls variants. In this work, we present an updated version of MycoSNP using Nextflow DSL2, making installation on UNIX systems easy and results highly reproducible across runs. In addition, new workflows add functionalities like phylogenetic tree-building and gene variant annotations that provide decision-making information to health scientists much faster. Wherever possible, new modules created for MycoSNP have been submitted to nf-core to make them available to all nf-core pipelines.

Hunter Seabolt
Bioinformatics Lead Scientist at the Centers for Disease Control and Prevention