#20nf-core/sarek: A workflow for germline, tumor-only, and somatic analysis of NGS data
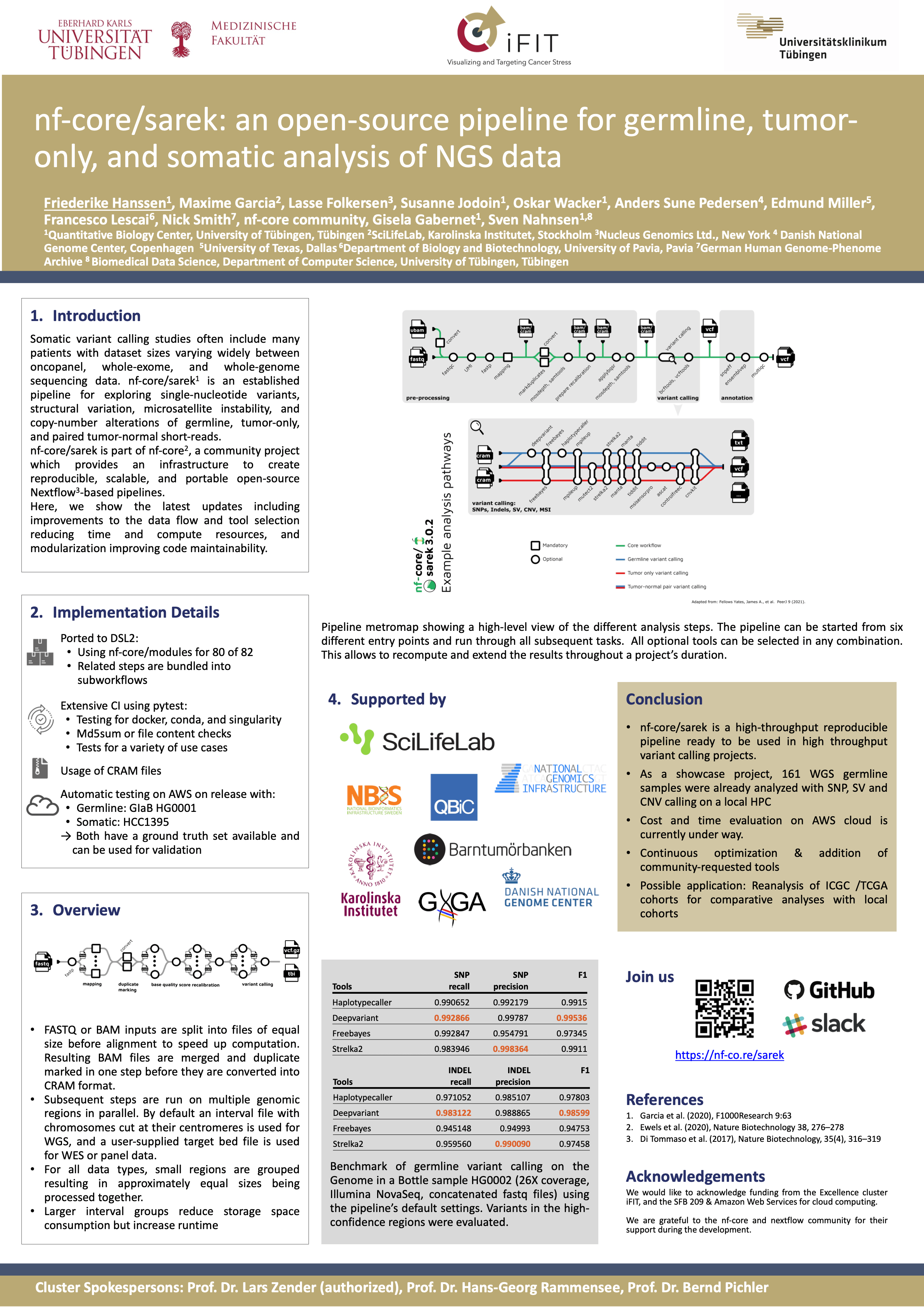
High-throughput, efficient, and reproducible pipelines are needed to ensure homogeneous data processing across different compute infrastructures with affordable resource usage. We present nf-core/sarek 3.0, to explore single-nucleotide variants, structural variation, microsatellite instability, and copy-number alterations of germline, tumor-only, and tumor-normal pairs.
We reduced compute resources and increased turn-around times, which minimizes costs on commercial clouds, facilitating the integration of publicly hosted data from repositories with in-house patient cohorts.
Other improvements include modularization of processes which facilitates maintainability and customization, and a broader repertoire of available tools.
We have re-processed 54 whole-genome-sequenced tumor-normal pairs of the TCGA-LIHC cohort, as well as on-site data, including 100 cholangiocarcinoma and 20 colorectal carcinoma panels to investigate the relationship of genomic variation to drug responsiveness.

Friederike Hanssen
Ph.D. Student at the Quantitative Biology Center, University of Tübingen