#6Efficient parallel taxonomic profiling for metagenomics with nf-core/taxprofiler
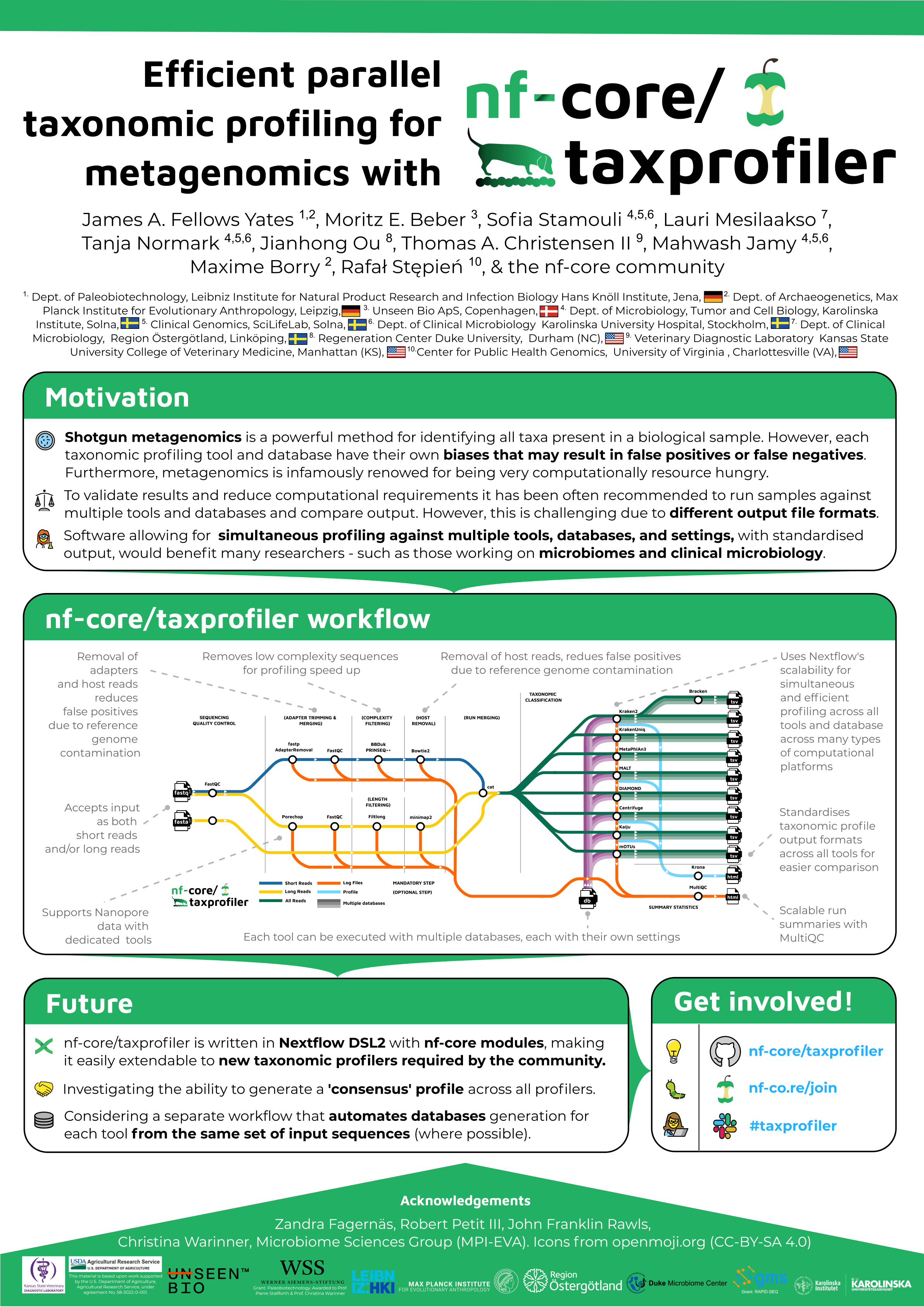
Taxonomic profiling addresses the challenge of genetically identifying all possible taxa that may be present in a metagenomic sample. However, different algorithms, parameters, and reference databases often yield different results. Selecting one tool or database risks per-tool or per-database biases, which might result in both false-negative and false-positive taxonomic classification.
nf-core/taxprofiler leverages Nextflow’s highly-resource efficient and parallelised job distribution to provide users simultaneous classification of both short and long read sequencing data across multiple metagenomic profiling tools and databases. The pipeline includes steps important for metagenomic researchers, involving removal of various sources of contamination, as well as standardisation of classification tables across all profilers. Written in Nextflow’s DSL2 and using nf-core modules, the pipeline is well-suited for continued integration of additional metagenomic profilers.
James Fellows Yates
Postdoctoral Bioinformatician at the Leibniz Institute for Natural Product Research and Infection Biology and Max Planck Institute for Evolutionary Anthropology